Part 1 of my series of posts on building prediction intervals used data held-out from model training to evaluate the characteristics of prediction intervals. In this post I will use hold-out data to estimate the width of the prediction intervals directly. Doing such can provide more reasonable and flexible intervals compared to analytic approaches1.
This post is not required for understanding part three of the series: Quantile Regression Forests for Prediction Intervals. However this post assumes you have already read Part 1, Understanding Prediction Intervals2:
- I do not re-explain terms such as coverage and interval width (or relative measures) – which are discussed thoroughly in the section on Reviewing Prediction Intervals.
- I do not reintroduce measures, figures, or chart types that were shown in the prior post.
- I continue using Ames, Iowa home sale prices as my example dataset.
This post was largely inspired by topics discussed in the Prediction intervals with tidymodels, best practices? Rstudio Community thread.
Update
I copied most of the code referenced at gists in this post to the spin package. The workboots package by Mark Rieke is essentially the same thing3 but is available on CRAN. I also created a placeholder spinach where I may do some future work…
{spinach} – Simulating Prediction INtervals And CHance – a #tidymodels #tidyverse friendly📦for a general interface to simulating uncertainty of predictions.
— Bryan Shalloway (@brshallo) April 1, 2022
Only a placeholder with a link to a prototype right now but hoping to start on soon: https://t.co/YpipdO5f1u #rstats pic.twitter.com/de2A4Z6WiD
Rough Idea
Compared to analytic approaches, estimating sources of uncertainty through simulation has the advantage of…
- relying on fewer model assumptions to still produce reasonable measures of uncertainty (though still generally requires that your model’s errors are iid).
- can be applied to (essentially4) any model type5.
Approach:
The simulation based technique I walk through will take resamples of the data and generate separate distributions for what can be loosely thought of as:
- the uncertainty due to model estimation
- the uncertainty due to the sample
These will then be combined to produce an overall measure of uncertainty for the prediction6.
Inspiration
Dan Saattrup Nielsen also wrote a series of posts on prediction intervals. My approach here is mostly taken from the one he describes in Boostrapping prediction intervals. His approach is encoded in python whereas mine is in R for a tidymodels based set-up7. (In the Appendix, I describe and link to an implementation of an Alternative Procedure With CV that is influenced by suggestions from Max Kuhn.)
Dan’s post provides more precise symbolic representations as well as more figures showcasing results and advantages of simulation based approaches, e.g.

Example of more sensible handling of non-normality of errors, from Dan Saattrup’s excellent post Bootstrapping prediction intervals
The Appendix provides links from the web to Other Examples Using Simulation.
Procedure
Given the model training dataset with n number of observations…
- Build b number of models using b bootstrap resamples on the training dataset (default of b is the square root of n)8.
- Use the 0.632+ rule for blending errors from analysis and assessment sets9 (based on amount of overfitting in analysis set) and create a distribution of residuals evenly spaced across quantiles of length n – represents distribution for the uncertainty due to the sample.
Given an observation (or set of observations) you would like to produce prediction intervals for…
- For each new observation, produce a prediction using all of the b models created in step 1.
- Take the difference between each model’s prediction (from step 3) and the mean of all model predictions – the resulting b differences for each observation provides the distribution for the variability due to model estimation at each point.
- For each observation, repeatedly sample from “residuals” (step 2) and “model error / differences” (step 4) and add together – do this h times (with replacement) OR create all possible combinations from step 2 and step 5. Rather than h, the latter approach will produce a distribution composed of n x b elements10 for each new observation (default behavior11).
- Pull quantiles (from the distribution created in step 5) according to the desired level of coverage of your intervals – e.g. 0.05 and 0.95 for a 90% prediction interval.
See gist for documentation on implementation.
Example
The initial set-up (load packages, load data, set pre-processing recipe, model specification) is the same as in the Providing More Than Point Estimates section of part 1. The code below is being sourced and printed from that post’s .Rmd file.
Load packages:
library(tidyverse)
library(tidymodels)
library(AmesHousing)
library(gt)
# function copied from here:
# https://github.com/rstudio/gt/issues/613#issuecomment-772072490
# (simpler solution should be implemented in future versions of {gt})
fmt_if_number <- function(..., digits = 2) {
input <- c(...)
fmt <- paste0("%.", digits, "f")
if (is.numeric(input)) return(sprintf(fmt, input))
return(input)
}Load data:
ames <- make_ames() %>%
mutate(Years_Old = Year_Sold - Year_Built,
Years_Old = ifelse(Years_Old < 0, 0, Years_Old))
set.seed(4595)
data_split <- initial_split(ames, strata = "Sale_Price", p = 0.75)
ames_train <- training(data_split)
ames_holdout <- testing(data_split) Specify pre-processing steps and model:
lm_recipe <-
recipe(
Sale_Price ~ Lot_Area + Neighborhood + Years_Old + Gr_Liv_Area + Overall_Qual + Total_Bsmt_SF + Garage_Area,
data = ames_train
) %>%
step_log(Sale_Price, base = 10) %>%
step_log(Lot_Area, Gr_Liv_Area, base = 10) %>%
step_log(Total_Bsmt_SF, Garage_Area, base = 10, offset = 1) %>%
step_novel(Neighborhood, Overall_Qual) %>%
step_other(Neighborhood, Overall_Qual, threshold = 50) %>%
step_dummy(Neighborhood, Overall_Qual) %>%
step_interact(terms = ~contains("Neighborhood")*Lot_Area)
lm_mod <- linear_reg() %>%
set_engine(engine = "lm") %>%
set_mode("regression")Specify a workflow, however do not yet fit the model to a training dataset.
lm_rec_mod <- lm_wf <- workflows::workflow() %>%
add_model(lm_mod) %>%
add_recipe(lm_recipe)Simulate Prediction Interval
Steps 1 & 2 from Procedure:
prep_interval() takes in a workflow (model specification + pre-processing recipe12) along with a training dataset, and outputs a named list containing bootstrapped model fits + prepped recipes (model_uncertainty) and the resulting residuals from cross-validation (sample_uncertainty).
# load custom functions `prep_interval()` and ``predict_interval()`
devtools::source_gist("https://gist.github.com/brshallo/4053df78265ab9d77f753d95f5faaf5b")
set.seed(1234)
prepped_for_interval <- prep_interval(lm_rec_mod, ames_train, n_boot = 200)
prepped_for_interval ## $model_uncertainty
## # A tibble: 200 x 2
## fit recipe
## <list> <list>
## 1 <fit[+]> <recipe>
## 2 <fit[+]> <recipe>
## 3 <fit[+]> <recipe>
## 4 <fit[+]> <recipe>
## 5 <fit[+]> <recipe>
## 6 <fit[+]> <recipe>
## 7 <fit[+]> <recipe>
## 8 <fit[+]> <recipe>
## 9 <fit[+]> <recipe>
## 10 <fit[+]> <recipe>
## # ... with 190 more rows
##
## $sample_uncertainty
## # A tibble: 2,197 x 1
## .resid
## <dbl>
## 1 -0.347
## 2 -0.321
## 3 -0.304
## 4 -0.291
## 5 -0.278
## 6 -0.266
## 7 -0.257
## 8 -0.247
## 9 -0.239
## 10 -0.232
## # ... with 2,187 more rowsSteps 3-6 in Procedure:
predict_interval() takes in the output from prep_interval(), along with the dataset for which we want to produce predictions (as well as the quantiles associated with our confidence level of interest), and returns a dataframe containing the specified prediction intervals.
pred_interval <- predict_interval(prepped_for_interval, ames_holdout, probs = c(0.05, 0.50, 0.95))
lm_sim_intervals <- pred_interval %>%
mutate(across(contains("probs"), ~10^.x)) %>%
bind_cols(ames_holdout) %>%
select(Sale_Price, contains("probs"), Lot_Area, Neighborhood, Years_Old, Gr_Liv_Area, Overall_Qual, Total_Bsmt_SF, Garage_Area)
lm_sim_intervals <- lm_sim_intervals %>%
rename(.pred = probs_0.50, .pred_lower = probs_0.05, .pred_upper = probs_0.95) %>%
relocate(c(.pred_lower, .pred_upper, .pred)) See gist for more documentation on prep_interval() and predict_interval().
Review
Reviewing our example offer from the Part 1 post, we see a roughly similar prediction interval to that specified by the analytic method:
lm_sim_intervals %>%
select(contains(".pred")) %>%
slice(1) %>%
gt::gt() %>%
gt::fmt_number(c(".pred", ".pred_lower", ".pred_upper"), decimals = 0)| .pred_lower | .pred_upper | .pred |
|---|---|---|
| 136,856 | 249,534 | 183,480 |
Though when reviewing a sample of observations, we notice an important contrast:
set.seed(1234)
lm_sim_intervals %>%
mutate(pred_interval = ggplot2::cut_number(Sale_Price, 10)) %>%
group_by(pred_interval) %>%
sample_n(2) %>%
ggplot(aes(x = .pred))+
geom_point(aes(y = .pred, color = "prediction interval"))+
geom_errorbar(aes(ymin = .pred_lower, ymax = .pred_upper, color = "prediction interval"))+
geom_point(aes(y = Sale_Price, color = "actuals"))+
labs(title = "90% prediction intervals on a holdout dataset",
subtitle = "Linear model (simulation method)",
y = "Sale_Price prediction intervals and actuals")+
theme_bw()+
coord_fixed()+
scale_x_log10(labels = scales::dollar)+
scale_y_log10(labels = scales::dollar)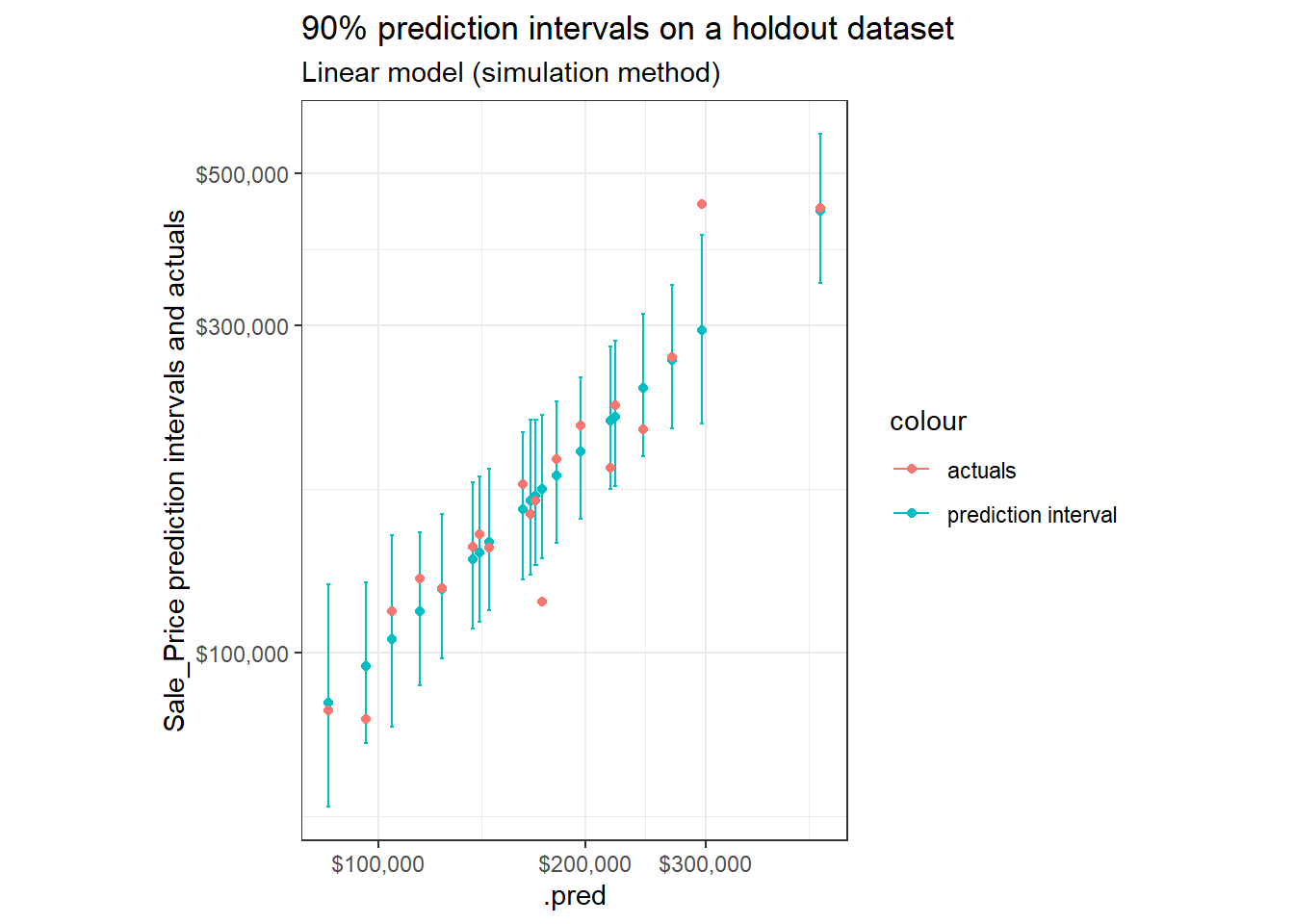
Simulation produced intervals show more differences in relative interval widths compared to those outputted with analytic methods (see Part 1, Review Prediction Intervals where the relative interval widths were nearly constant across predictions).
Interval Width
The relative widths of prediction intervals from the simulation based approach vary between observations by more than 10x what we saw in the Interval Widths using analytic methods13 (where interval width had a standard deviation of less than half a percentage point).
We also see greater differences in interval width between buckets of predictions (a range of about 5 to 6 percentage points vs 0.4).
This suggests the simulation based approach allows for greater differentiation in estimated levels of uncertainty by attributes14.
lm_sim_widths <- lm_sim_intervals %>%
mutate(interval_width = .pred_upper - .pred_lower,
interval_pred_ratio = interval_width / .pred) %>%
mutate(price_grouped = ggplot2::cut_number(.pred, 5)) %>%
group_by(price_grouped) %>%
summarise(n = n(),
mean_interval_width_percentage = mean(interval_pred_ratio),
stdev = sd(interval_pred_ratio),
stderror = sd(interval_pred_ratio) / sqrt(n)) %>%
mutate(x_tmp = str_sub(price_grouped, 2, -2)) %>%
separate(x_tmp, c("min", "max"), sep = ",") %>%
mutate(across(c(min, max), as.double)) %>%
select(-price_grouped)
lm_sim_widths %>%
mutate(across(c(mean_interval_width_percentage, stdev, stderror), ~.x*100)) %>%
gt::gt() %>%
gt::fmt_number(c("stdev", "stderror"), decimals = 2) %>%
gt::fmt_number("mean_interval_width_percentage", decimals = 1)| n | mean_interval_width_percentage | stdev | stderror | min | max |
|---|---|---|---|---|---|
| 147 | 55.7 | 8.44 | 0.70 | 43200 | 121000 |
| 146 | 51.0 | 4.97 | 0.41 | 121000 | 145000 |
| 146 | 50.2 | 2.84 | 0.23 | 145000 | 176000 |
| 146 | 50.7 | 5.07 | 0.42 | 176000 | 221000 |
| 146 | 51.8 | 8.02 | 0.66 | 221000 | 485000 |
Simultaneously, the average relative interval width is slightly more narrow: about 52% for the simulation based approach against 54% for the analytic method15.
lm_sim_intervals %>%
mutate(interval_width = .pred_upper - .pred_lower,
interval_pred_ratio = interval_width / .pred) %>%
summarise(n = n(),
mean_interval_width_percentage = mean(interval_pred_ratio),
stderror = sd(interval_pred_ratio) / sqrt(n)) %>%
mutate(across(c(mean_interval_width_percentage, stderror), ~.x * 100)) %>%
gt::gt() %>%
gt::fmt_number(c("mean_interval_width_percentage", "stderror"), decimals = 2) %>%
gt::fmt_number("mean_interval_width_percentage", decimals = 1)| n | mean_interval_width_percentage | stderror |
|---|---|---|
| 731 | 51.9 | 0.24 |
Coverage
Coverage is essentially the same: 92.2% (vs 92.7% for Coverage with the analytic method)16.
lm_sim_intervals %>%
mutate(covered = ifelse(Sale_Price >= .pred_lower & Sale_Price <= .pred_upper, 1, 0)) %>%
summarise(n = n(),
n_covered = sum(
covered
),
stderror = sd(covered) / sqrt(n),
coverage_prop = n_covered / n) %>%
mutate(across(c(coverage_prop, stderror), ~.x * 100)) %>%
gt::gt() %>%
gt::fmt_number("stderror", decimals = 2) %>%
gt::fmt_number("coverage_prop", decimals = 1)| n | n_covered | stderror | coverage_prop |
|---|---|---|---|
| 731 | 674 | 0.99 | 92.2 |
Coverage rates across quintiles:
lm_sim_intervals %>%
mutate(price_grouped = ggplot2::cut_number(.pred, 5)) %>%
mutate(covered = ifelse(Sale_Price >= .pred_lower & Sale_Price <= .pred_upper, 1, 0)) %>%
group_by(price_grouped) %>%
summarise(n = n(),
n_covered = sum(
covered
),
stderror = sd(covered) / sqrt(n),
n_prop = n_covered / n) %>%
mutate(x_tmp = str_sub(price_grouped, 2, -2)) %>%
separate(x_tmp, c("min", "max"), sep = ",") %>%
mutate(across(c(min, max), as.double)) %>%
ggplot(aes(x = forcats::fct_reorder(scales::dollar(max), max), y = n_prop))+
geom_line(aes(group = 1))+
geom_errorbar(aes(ymin = n_prop - 2 * stderror, ymax = n_prop + 2 * stderror))+
coord_cartesian(ylim = c(0.70, 1.01))+
# scale_x_discrete(guide = guide_axis(n.dodge = 2))+
labs(x = "Max Predicted Price for Quintile",
y = "Coverage at Quintile",
title = "Coverage by Quintile of Predictions",
subtitle = "On a holdout Set",
caption = "Error bars represent {coverage} +/- 2 * {coverage standard error}")+
theme_bw()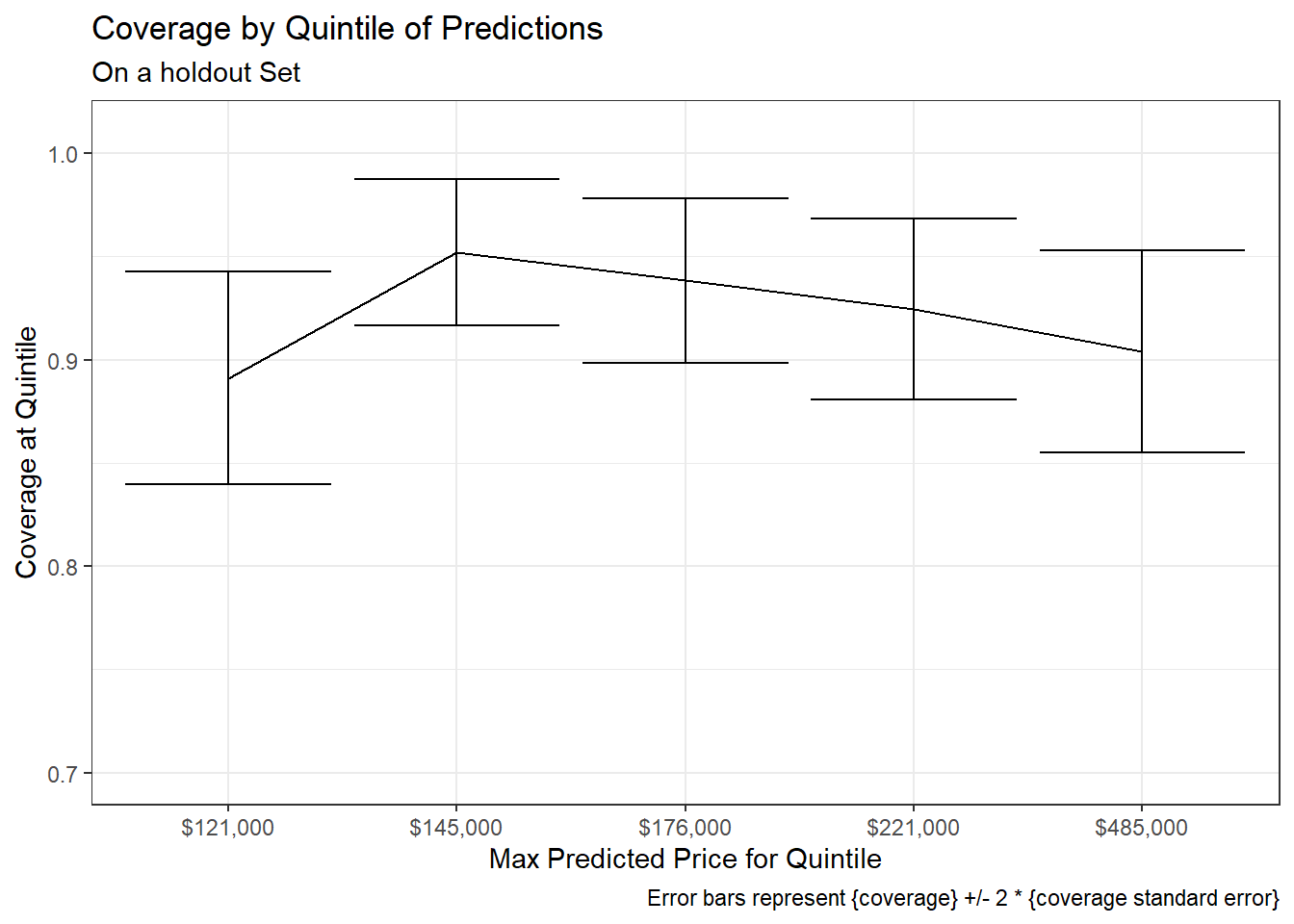
The overall pattern appears broadly similar to that seen in the analytic method. However coverage rates seem to be slightly more consistent across quintiles in the simulation based approach – an improvement over the analytic method (where coverage levels had appeared to be slightly more dependent upon quintile of predicted Sale_Price)17.
For example, a chi-squared test in the previous post had shown significant variation in coverage rates across quintiles when applied to the analytic method. However the same test applied to the results from the simulation based prediction intervals does not show a significant difference18.
lm_sim_intervals %>%
mutate(price_grouped = ggplot2::cut_number(.pred, 5)) %>%
mutate(covered = ifelse(Sale_Price >= .pred_lower & Sale_Price <= .pred_upper, 1, 0)) %>%
with(chisq.test(price_grouped, covered)) %>%
pander::pander()| Test statistic | df | P value |
|---|---|---|
| 4.987 | 4 | 0.2886 |
Keep in mind we are looking at just one model specification on one set of data19.
See Adjusting Procedure in the Appendix for a couple examples of circumstances where you might want to make changes to Procedure.
Closing Notes
This post walked through a rough implementation for simulating prediction intervals for arbitrary model types + preprocessing steps in a way that is loosely in the style of tidymodels. It represents a continuation on Understanding Prediction Intervals and is the second of three posts I am writing on prediction intervals. The third post will cover quantile regression.
Advantages of simulation based techniques for building prediction intervals:
- can produce prediction intervals for model types that are intractable to produce estimates for analytically.
- Even for model types that have analytic solutions for prediction intervals, simulation based approaches rely on fewer assumptions and can be more flexible20.
Downsides:
- These methods are far more computationally expensive as you will need to build a lot of models21.
- See Tidy Models with R, 10.4 for notes on how to set-up Parallel Processing.
- For predictive inference, you still can’t drop all assumptions22.
Appendix
Conformal Inference
Highly related to what I presented in this post, the academic discipline of using out-of-sample data to create prediction intervals is known as conformal inference (much of the research in this field comes from Carnegie Mellon University and Royal Holloway University, London). I may do a follow-up post where I walk through a more formal example of using conformal inference. In the meantime, here are a few resources I glanced through23:
- ryantibs/conformal: github repo with
conformalInferenceR package and links to relevant articles on distribution-free predictive inference. The way model types are specified byconformalInfereneseems to be not too dissimilar from the approach I took in this post24 - donlnz/nonconformist: python package for conformal inference
- Conformal Prediction: Link to Royal Holloway University website by creators of method – Vladimir Vovk and Alex Gammerman.
- Assumption-free prediction intervals for black-box regression algorithms - Aaditya Ramdas (YouTube): professor at CMU giving overview of problem, approaches, and current “state-of-the-art”.
- Tutorial on conformal inference, Dataiku article, Analytics Vidhya article
Other Examples Using Simulation
Here is a video walk-through of a simple example using a linear model:
A slightly modified version of this approach that adjusts for the leverage of the observations (leverage25 has to do with distance of a point from the centroid of the data26) can be found at this Cross Validated Thread.
Limitation with these examples (as encoded):
- re-estimate some component of their prediction intervals based on performance on the data used to train the model.
- are set-up for linear models
Confusion With Confidence Intervals
It seems to be common for people to seem to set-out to simulate prediction intervals but end-up simulating confidence intervals27 – i.e. they account for uncertainty in estimating the model while forgetting to account for the uncertainty of the sample. I wrote more on this in Prediction Intervals and Confidence Intervals from Part 128.
Adjusting Procedure
Below are just a few examples. (Procedure is more just a toy set-up and by no means optimized.)
Coverage level depends on attribute(s)
If we see that coverage rates depend on predicted value of Sale_Price (or on some other attribute) we might:
- try altering the pre-processing recipe
- adjust Procedure to segment29 the residuals by predicted
Sale_Priceand shuffle the errors within segments (rather than across the entire dataset). This would allow the uncertainty due to the sample to vary according to the predictedSale_Price30.
Random Splits Not Appropriate
The procedure I walk through assumes a random split in your data is fine and that your observations are iid. This is generally a requirement for using the bootstrap, however there are some approaches to using modified bootstrap that can adjust for this. In many cases it may make sense to have group or time based sampling schemes (see my presentation that discusses time-based resampling schemes). If a random sampling scheme is inappropriate, you will likely end-up creating too small of prediction intervals. I had considered making prep_interval() capable of taking in custom resampling specifications but did not set this up… but could be potentially edited.
Alternative Procedure With CV
Given the model training dataset with n number of observations…
- Build b number of models using b bootstrap resamples on the training dataset (default of b is the square root of n)31.
- Build another set of k models32 using k-fold cross-validation33 (default of k is 10) and extract the residuals, composed of n elements – providing distribution for uncertainty due to the sample34.
Given an observation (or set of observations) you would like to produce prediction intervals for…
- For each new observation, produce a prediction using all of the b models created in step 1.
- Take the difference between each model’s prediction (from step 3) and the mean of all the model’s predictions – the resulting b differences for each observation provides the distribution for the variability due to model estimation at each point.
- For each observation, repeatedly sample from “residuals” (step 2) and “model error / differences” (step 4) and add together – do this h times (with replacement) OR create all possible combinations from step 2 and step 5. Rather than h, the latter approach will produce a distribution composed of n x b elements35 for each new observation (default behavior36).
- Pull quantiles (from the distribution created in step 5) according to the desired level of coverage of your intervals – e.g. 0.05 and 0.95 for a 90% prediction interval.
See gist for documentation on implementation of this method.
Differences from Procedure
This approach is influenced by suggestions from Max Kuhn. For example, I …
- only use out-of-sample estimates to produce the interval
- estimate the uncertainty of the sample using the residuals from a separate set of models built with cross-validation37
For example when the model assumptions associated with analytic methods are broken.↩︎
This post should be viewed as simply another section of Understanding Prediction Intervals rather than as entirely self-contained. It is essentially a “How To” for simulating prediction intervals with tidymodels.↩︎
As of now. Mark may be making some additions in the future though per chats at Twitter thread.↩︎
With the caveat that some model types may require lots of simulations for the interval produced to be appropriate as discussed here by Max Kuhn. Also, there is usually the assumption that errors are exchangable across observations.↩︎
The trade-off with simulation techniques is generally you get to dodge hairy or intractable math at the cost of high computation costs.↩︎
I elaborated on intuitions for these sources of uncertainty in a A Few Things to Know About Prediction Intervals↩︎
Mine also includes supporting a pre-processing recipe in addition to a model specification.↩︎
Per guidance on Nielsen’s post – though Kuhn suggests [here]((https://community.rstudio.com/t/prediction-intervals-with-tidymodels-best-practices/82594/4?u=brshallo) that more samples should be taken.↩︎
The reason this is done is that the errors on the analysis set may be too small and on the assessment set too big (because are not using full dataset to train on any one model) so this tries to find a balance between these.↩︎
{number of observations in model training dataset} x {number of models created}↩︎
Warning that this will become computationally expensive very quickly as number of observations in training data or number of models created increases.↩︎
Because a workflow contains both a function for fitting models and a pre-processing recipe, this procedure accounts for variability due to pre-processing as a part of the model-fitting procedure.↩︎
This is a rough calculation, but I’m just looking at the standard deviations in the two respective tables. For the lowest bucket for example, the analytic method had a standard deviation of 0.46 percentage points, for the simulation based technique it is 8.44 percentage points, 8.44 / 0.46 which is closer to 20x, but the difference is not quite so big in other groups.↩︎
E.g. based on the attributes of an observation. Remember that the analytic method only allowed for slight variability in prediction intervals across observations. Based on a constrained definition of the distance of an observation from the centroid of the data.↩︎
This suggests a slightly more precise accounting of uncertainty.↩︎
+/- ~2 percentage points↩︎
The reason you want consistent coverage rates is because then you can trust your interval captures your data at the rate you would expect across observations and not just in aggregate. The difference is related to conditional vs marginal coverage.↩︎
There are more direct ways available for comparing these. This, after all, isn’t a test comparing the two distributions but two separate tests against the NULL hypothesis. Also noted in a footnote in Part 1, it would likely have been better to base the groupings on
Sale_Pricerather than predictedSale_Priceso could have had paired groups. Rather than a general chi-squared test, something like McNemar’s test may be more appropriate.↩︎Further examples would be needed to make more general statements on the qualities of simulation versus parametric based approaches to building prediction intervals.↩︎
For example, you generally don’t need to worry about having “normality of errors” – simulation will generate whatever distribution your errors follow.↩︎
If Procedure was changed could make less expensive in some cases. E.g. Some linear model types have computational tricks available that can make this far less taxing. Some out-of-sample techniques do not actually require more data at all. Some related techniques (e.g. split-conformal) may only require a few more models.↩︎
See The limits of distribution-free conditional predictive inference, Barber, et al.↩︎
In that it takes in a model generating algorithm as input – seems could set-up interface or something similar in a way that is pretty tidy friendly (e.g.
add_conformal()…↩︎And is usually thought of in the context of linear regression.↩︎
The influence of observation centrality was discussed extensively in Part 1 – points further from the center of the data have greater uncertainty in model estimation.↩︎
Example from response on Rstudio Community thread. Or else they are ambiguous about which they intend to build as in this example from an online textbook on Inferential Thinking.↩︎
This confusion may be particularly common in the case of simulation because bootstrapping is generally concerned with estimating the distribution of parameter estimates rather than individual observations.↩︎
Or perhaps segmented in a fuzzy way so rather than having discrete bins is done on a potentially continuous scale↩︎
Or any other selected attribute – though would need to put some more thought into how to do properly, appropriate number of observations, etc., hard or fuzzy segmenting, how many buckets or how to handle continuously…↩︎
Per guidance on Nielsen’s post – though Kuhn suggests here that more samples should be taken.↩︎
Though functions can also be set such that will just use the bootstrapped models (rather than building a separate set).↩︎
I think Max suggested using cross-validation here instead of the residuals on the out-of-bag samples in the bootstraps because bootstrap samples tend to overestimate the errors (I believe).↩︎
I ignore any adjustments due to centroid as I’m not sure how to do this appropriately outside of the linear regression context.↩︎
{number of observations in model training dataset} x {number of models created}↩︎
Warning that this will become computationally expensive very quickly as number of observations in training data or number of models created increases.↩︎
Rather than the residuals of the bootstrapped samples used to estimate the uncertainty of the model. (However the functions I create do allow for using the bootstrapped resamples to calculate the uncertainty in the sample – see gist)↩︎